
StudySmarter - The all-in-one study app.
4.8 • +11k Ratings
More than 3 Million Downloads
Free
Americas
Europe
How do our cells go from being a zygote cell to becoming other types of cells like muscle cells, red blood cells, or even osteoblasts? Regulation of gene expression and cell differentiation takes place! Chromatin modification is one of the ways in which regulation of gene expression occurs. Interested in learning more about how chromatin is modified? Keep reading!
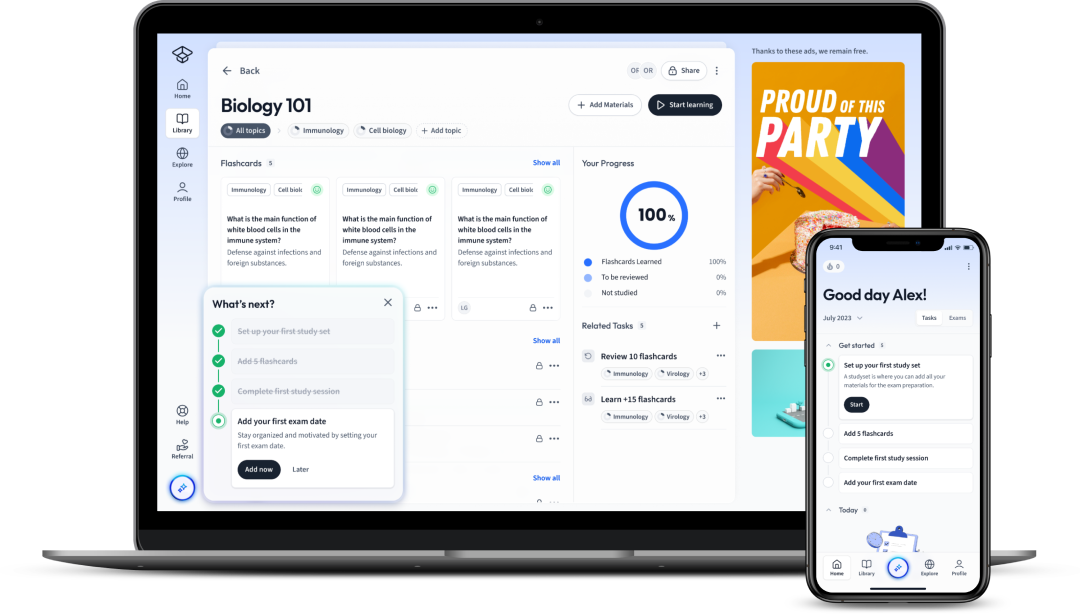
Explore our app and discover over 50 million learning materials for free.


























Lerne mit deinen Freunden und bleibe auf dem richtigen Kurs mit deinen persönlichen Lernstatistiken
Jetzt kostenlos anmeldenHow do our cells go from being a zygote cell to becoming other types of cells like muscle cells, red blood cells, or even osteoblasts? Regulation of gene expression and cell differentiation takes place! Chromatin modification is one of the ways in which regulation of gene expression occurs. Interested in learning more about how chromatin is modified? Keep reading!
To be able to understand what chromatic modification really means, we need to be familiar with the concept of differential gene expression.
The human body is made up of more than 200 types of cells, and all of these cells are genetically identical, meaning that they have the same genome/DNA. But, each cell has a different proteome (set of proteins). So, differences between cell types (cellular differentiation) are simply a result of differential gene expression.
For example, liver cells and skin cells are genetically identical, but different genes are expressed.
Regulation (control) of gene expression is very important for cell specialization (also known as cell differentiation) because it allows organisms to be more efficient with resources and adapt to the environment.
Regulation of eukaryotic gene expression occurs at any of the following stages:
Chromatin rearrangement/Chromatin modification: regulates chromatin conformation and accessibility of DNA for transcription.
Transcriptional control: the regulation of RNA polymerase binding to a promoter as a way to regulate/control transcription.
Post-transcriptional control: regulates modifications to RNA after transcription.
Translational control: regulates the steps of initiation and elongation in translation.
Post-translational control: regulates modifications to proteins after translation. Often, the final product for the regulation of eukaryotic gene expression is a protein!
Remember: gene expression involves of the processes of transcription and translation (transforming DNA into RNA and then RNA into proteins! For an in-depth explanation of all steps in the regulation of eukaryotic gene expression, check out "Eukaryotic Gene Expression"!
Chromatin modification is the first step in the regulation of eukaryotic gene expression, and it happens before transcription. As the name suggests, chromatin modification is the modification of the chromatin structure of eukaryotes for regulating their gene expression.
Remember that DNA in eukaryotic cells is packaged with proteins in an elaborate complex called chromatin.
Chromatin is loosely packed/coiled nucleosomes (DNA wrapped around units of 8 histone proteins, repeatedly).
DNA has a negative charge due to phosphate groups, whereas histones carry a positive charge from lysine (amino acid Lys) residues. So, this attraction between opposite charges is what makes the DNA to wrap around histones and form chromatin.
Chromatin is found inside the nucleus of a eukaryotic cell.
Chromatin modification is the process of modifying histone proteins or DNA sequences to control transcription.
To learn about gene expression in prokaryotes, read "Prokaryotic Gene Expression"!
There are two different types of chromatin, based on DNA compaction in the chromosome: heterochromatin and euchromatin.
Heterochromatin is the condensed region of the genome with low transcriptional activity.
Euchromatin is the loosely packed region of the genome, with high transcriptional activity and histone/DNA modifications.
When we talk about chromatin modification, we are most likely talking about modifications that occur on histones and DNA. The main function of chromatin modification is to prepare the chromatin for DNA replication and transcription, and this is achieved in two ways:
Disrupting the interaction of histones with DNA.
Affecting the recruitment of non-histone proteins (proteins with enzymatic activities) to modify chromatin.
There are different chromatin modifications identified on histones. These are acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation (small ubiquitin-like modifier proteins), ADP ribosylation, deimination, and proline isomerization. All of these modifications function as regulators of transcription, but some of them can also regulate repair and DNA replication.
Although scientists have found nine types of chromatin modifications in eukaryotic cells, we will only talk about three of them: histone acetylation, DNA methylation, and histone methylation.
One of the most common types of chromatin modification is histone acetylation. In this process, the chromatin structure loosens, making the DNA more accessible to transcription factors (proteins that regulate gene expression by directly binding to DNA), eventually resulting in the activation of RNA polymerase to bind DNA and start transcription!
Histones are proteins that contain a long polypeptide "tail" composed of 25-40 amino acids. This tail can be chemically modified by cellular enzymes.
Histone acetylation is the attachment of an acetyl group (-COCH3) to histone molecules to loosen their grip on the DNA molecule, forming euchromatin. Histone acetylation is associated with transcriptional activation.
Basically, in order to "free" the DNA from being wrapped around the histone, we need to get rid of the positive charge on the histone to stop the positively charged DNA from being attracted to it. We do this by attaching an acetyl group to the histone molecules with the help of an enzyme called histone acetyltransferase (HAT). This causes the histones to go from a positive to a neutral charge, decreasing histone affinity for DNA.
Now that DNA is unpackaged it is ready for the transcription!
Histone acetylation is reversible. The opposite process, which is removing the acetyl group from the histone, is called de-acetylation and involves the enzyme histone deacetylase (HDAC). De-acetylation leads to tight packing of the chromatin structure (heterochromatin), and is associated with gene silencing.
DNA methylation is a type of chromatin modification that prevents/"turns off" DNA transcription by blocking the access of transcription factors to the DNA (ex: blocks RNA polymerase's access to the promoter).
DNA methylation is the addition of a methyl group (-CH3) to the cytosine nucleotides of DNA, modifying the DNA sequence and blocking transcription. The enzymes that drive this process are called DNA methyl transferases (DNMTs).
How does DNA methylation happen? During DNA methylation, a methyl group gets attached to one of the carbon atoms in the cytosine molecule, turning cytosine into S-methyl cytosine.
This modification in the DNA sequence prevents transcription from happening.
Methylation can also happen on histones. During histone methylation, a methyl group is added to the histone. Histone methylation leads to better access of transcription factors to DNA, and the activation of gene transcription if a single methyl group is added. But, If 2-3 methyl groups are attached instead, the histone tails will tighten, reducing access for transcription factors and blocking transcription!
Another type of chromatin modification is X-inactivation, the process of creating an entirely inactivated X-chromosome in females. X-inactivation occurs after fertilization when one of the two X sex chromosomes in females gets inactivated by the loss of acetylation in the histone proteins of nucleosomes. This inactivation is a way of equalizing the gene dosage that both males (who only have one X chromosome) and females express!
As an example, let's take a look at how cancer and chromatin modifications are related. In some cases, the mutation of regulatory genes that affect DNA methylation and histone acetylation leads to the development of cancer cells (tumors).
For example, a mutation of the DNMT3A regulatory gene in DNA methylation can cause acute myeloid leukemia. Other cancers associated with DNMT3A mutations are lung adenocarcinoma and colon adenocarcinoma.
A mutation in tumor suppressor gene MLL2 affects histone methylation and might lead to follicular lymphoma, whereas a mutation in gene MLL1 affects histone methylation and causes acute leukemia in infants.
Did you know that chromatin modification can also affect fungi? Researchers found that in plant pathogenic fungi such as Ustilago maydis (rust fungus), chromatin modification helps control host interaction and their virulence capacity!
In chromatin modification, histone proteins or DNA gets modified as a way of controlling transcription.
Chromatin modifiers are non histone proteins that attach to histones of DNA bases to activate or repress transcription.
Chromatin modification occurs in the nucleus of eukaryotic cells.
Chromatin modifications control gene expression by Disrupting the interaction of histones with DNA and also by affecting the recruitment of non-histone proteins (proteins with enzymatic activities) to modify chromatin.
In some cases, the mutation of regulatory genes that affect DNA methylation and histone acetylation lead to the development of cancer cells (tumors).
Flashcards in Chromatin Modification15
Start learningTrue or false: All cells in the human body are genetically identical (they have the same genome).
True
Differences between cell types (liver cell vs. neuron cell vs. red blood cells) are due to ________.
differential gene expression.
_______ is a process that allows multicellular organisms to express genes differently in different cells.
Differential gene expression
______ is the first step in the regulation of eukaryotic gene expression.
Chromatin modification
Chromatin are loosely packed/coiled ________ (DNA wrapped around histone proteins)
nucleosomes
Already have an account? Log in
Open in AppThe first learning app that truly has everything you need to ace your exams in one place
Sign up to highlight and take notes. It’s 100% free.
Save explanations to your personalised space and access them anytime, anywhere!
Sign up with Email Sign up with AppleBy signing up, you agree to the Terms and Conditions and the Privacy Policy of StudySmarter.
Already have an account? Log in