
StudySmarter - The all-in-one study app.
4.8 • +11k Ratings
More than 3 Million Downloads
Free
Americas
Europe
In eukaryotes, one way to control gene expression is by epigenetic changes.
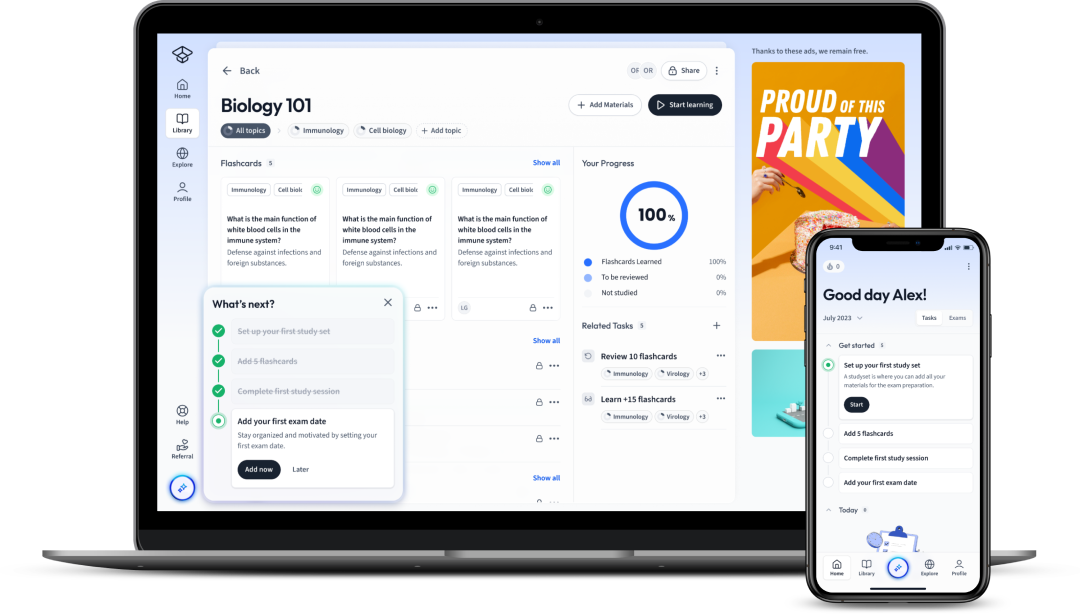
Explore our app and discover over 50 million learning materials for free.


























Lerne mit deinen Freunden und bleibe auf dem richtigen Kurs mit deinen persönlichen Lernstatistiken
Jetzt kostenlos anmeldenIn eukaryotes, one way to control gene expression is by epigenetic changes.
“Epigenetics is the study of mechanisms that lead to changes in gene expression that can be passed from cell to cell and are reversible, but do not involve a change in the sequence of DNA” 1
Epigenetic changes are heritable changes in gene expression that, unlike mutations, do not change the base sequence of DNA. These changes occur via the attachment or removal of tags (chemical groups) to the DNA or the histone proteins.
The long-term persistence of changes in gene expression is an important characteristic of epigenetic effects.
Let’s look at muscle cells in humans. Some genes in the human genome should be kept silenced in muscle cells since their expression would be wasteful or detrimental to the efficiency of muscle contraction. These genes are suppressed during embryonic development by epigenetic modifications such as DNA methylation that suppresses these genes.
During embryonic development, epigenetic modifications are transferred from cell to cell. For example, epigenetic changes in adult muscle cells prevent the expression of genes that are not needed for the function of the muscle since the expression would be wasteful.
Some epigenetic changes, such as the ones mentioned above, are relatively permanent during an individual’s lifespan, while others may be reversible during the individual’s life.
In multicellular organisms that undergo sexual reproduction, some epigenetic modifications may be passed from the parents to the offspring. This type of inheritance is called transgenerational epigenetic inheritance.
Environmental factors such as stress, toxins, and agents in cigarette smoke can result in epigenetic changes to DNA and affect gene expression.
The three most common chemical modifications involved in epigenetic regulation of gene expression are DNA methylation, chromatin remodelling, and covalent histone modification.
These changes can also occur in temporary/non-epigenetic control of gene expression.
The set of all epigenetic modifications (chemical group tags) on DNA is referred to as the epigenome.
One method that eukaryotes use to suppress gene expression is called DNA methylation.Methylation of DNA is when methyl groups (--CH3) are covalently attached to the carbon at the number 5 position of the cytosine base on the DNA by an enzyme called DNA methyltransferase, forming 5-methylcytosine. The methylated cytosine is usually connected to a neighbouring guanine residue via a phosphodiester bond on the same strand. These dinucleotides of cytosine and guanine are called CpG islands.
Methylation of the cytosine bases of CpG islands on both DNA strands is called full methylation, while methylation of cytosine bases only on one strand is termed hemimethylation.

DNA methylation usually inhibits the transcription of genes in eukaryotes, especially when it occurs near the gene’s promoter region. There are many CpG islands located near the promoter in both vertebrates and invertebrates.
In the housekeeping genes (essential genes for most cells’ viability in multicellular organisms, such as the gene for RNA polymerase gene), the CpG islands are mainly unmethylated. As a result, housekeeping is expressed in almost all cell types. On the other hand, some tissue-specific genes that are highly regulated may be suppressed by the methylation of CpG islands.
There are two ways by which methylation of CpG islands inhibits DNA transcription:
First, methylation may influence regulatory transcription factors (TFs) binding to the DNA. For instance, the presence of the methyl group may prevent the binding of an activatory TF to the enhancer region. This would inhibit the recruitment of RNA polymerase to the promoter and inhibit transcription initiation.
Second, methylation suppresses the expression of genes via proteins called methyl-CpG-binding proteins. These proteins can specifically recognise and bind to methylated CpG groups. After that, these proteins would recruit other proteins that inhibit transcription. For example, they may recruit a histone deacetylase to the methylated CpG region near the promoter, which would remove acetyl groups from the histones and make it more difficult for the histones to be removed from the DNA.
Chromatin is a DNA-protein complex within a chromosome. It’s how DNA presents itself while the cell is not going through cell division.
In eukaryotic cells, different areas within the chromatin can have different conformations. Areas of chromatin that are densely packed and have a closed structure are called heterochromatin. Due to its dense organisation, transcription of genes within the heterochromatin is very difficult or near impossible.
More open areas of the chromatin are known as euchromatin. Due to their open conformation, DNA polymerase and TFs can access euchromatin genes more easily than heterochromatin. As a result, the genes in euchromatin are expressed more than the genes located in heterochromatin.
The chromatin can be remodelled. One way to perform this is via ATP-dependent chromatin remodelling. This process is carried out by protein complexes that can recognise nucleosomes.
The simplest repeating unit of eukaryotic chromatin is called the nucleosome. A nucleosome consists of around 150 base pairs of DNA wrapped around 8 histone proteins.
These complexes contain a catalytic ATPase subunit called DNA translocase, which can use the energy released from ATP hydrolysis to change the position or composition of nucleosomes, which can make the chromatin more or less condensed.
Histone complexes are structures that DNA double helix is wrapped around in nucleosomes. Each of these complexes is composed of eight histone core proteins that consist of a globular domain and a flexible charged amino-terminal tail.
The amino-terminal tails protrude away from the chromatin while the DNA twists around the globular domains. The amino-terminal tails contain particular amino acids that are subject to various covalent modifications such as acetylation, methylation and phosphorylation.
Histone modification affects the interactions within the nucleosomes. Let’s take acetylation, for example. We briefly mentioned histone acetylation when we were discussing DNA methylation. During histone acetylation, an acetyl group (CH3CO-) is added to a positively charged lysine residue in the amino-terminal tail of a histone core protein by histone acetyltransferase. This attachment eliminates the positive charge on the lysine side chain, disrupting the electrostatic attraction between positively charged histones and negatively charged DNA strands. As a result, the DNA becomes less tightly bound to the histone proteins and becomes more accessible to RNA polymerase and TFs.

Histone acetylation is a reversible modification, and the acetyl group can be removed from the histone core proteins by the enzyme histone deacetylase. Decreased acetylation means lysine is in its positively charged state and has a stronger attraction to the DNA molecule, causing the association between DNA and histones to be stronger, so the DNA is not available to transcription factors. Therefore mRNA production is not initiated, and the gene is switched off - it cannot be transcribed or translated.
As discussed earlier, epigenetic effects result in DNA and chromosomal modifications that alter gene expression. Some of these epigenetic modifications may be reversible during an individual’s lifespan, but others may be permanent or even get passed on to the next generation. For instance, it has been shown that gestational diabetes in a pregnant mother can increase the likelihood of the daughter developing gestational diabetes in the future. These high glucose concentrations cause epigenetic changes in the daughter’s DNA.
Gestational diabetes is a condition in pregnant women that results in elevated levels of blood glucose levels in both the mother and the fetus.
Despite the cellular mechanism that searches and clears epigenetic changes in the DNA of sperm and egg cells, some modifications may escape and get passed on.
Dosage compensation describes the phenomenon which ensures the level of sex gene expression (such as those on the X chromosome) are the same in both males and females, even though males and females have different numbers of sex chromosomes.
Female mammals have two copies of the X chromosome, while males have only one. In this case, dosage compensation of sex genes occurs by X chromosome inactivation (XCI) in females.
XCI is mediated by epigenetic modifications that result in condensing one of the X chromosomes in the interphase nuclei of somatic cells in females, rendering the chromosome inactive. A short region of the X chromosome called the X inactivation centre (Xic) plays a critical role in X inactivation. If one of the two X chromosomes is missing its Xic due to a mutation, then the cell only recognizes one Xic, and as a result, X chromosome inactivation does not proceed. Having two active X chromosomes can be a lethal condition for a female human embryo!
The inactive X chromosome is known as the Barr body.
XCI occurs in three phases: initiation, spreading and maintenance.
Genomic imprinting refers to marking a segment of DNA where the mark is retained and recognized throughout the organism’s life, inheriting the marked DNA. Genomic imprinting occurs before fertilization and mainly involves a modification in a single gene or chromosome during gametogenesis.
Gametogenesis is how gametes, or germ cells, are produced in an organism.
Depending on whether the imprinting occurs during the sperm or the egg cell formation, each offspring expresses only one of the two alleles. This phenomenon is termed monoallelic expression.
Epigenetic modifications are a regulated process, forming part of healthy development. Changing any of the epigenetic mechanisms might result in aberrant gene activation or silencing and cause certain disorders. Such changes have been linked to various disorders, one of which is cancer.
As we discussed earlier, epigenetic changes do not affect the base sequence in the DNA. However, they can increase the rate of mutations.
Epigenetic changes are linked to cancer development. They are present in tumour cells and often cooperate with genetic alterations that drive the properties of cancer cells.
Tumour suppressor genes (TSGs) code for proteins that slow down cell division. They can be thought of as stop breaks in the cell cycle. TSGs can trigger cell death (apoptosis) if mistakes in the process of DNA replication are detected.
If a mutation occurs in TSGs, the protein produced would be non-functional. This would lead to uncontrolled cell division and unregulated proliferation. Examples of important TSGs in the cells are the p53 retinoblastoma genes. Other examples include the BRCA1 and BRCA2 genes linked to breast cancer development.
TSGs can become inactivated by three pathways:
Loss-of-function mutation.
Complete deletion.
Suppression by epigenetic changes.
Epigenetic silencing of TSGs can occur by deregulation of the machinery responsible for epigenetic modifications. For instance, it may involve inappropriate methylation of CpG islands within the promoter region of TSGs, thereby suppressing their transcription.
Proto-oncogenes are another set of genes that are important in the pathophysiology of tumours. Proto-oncogenes are normal genes that promote cell proliferation and can be thought of as gas pedals that accelerate the cell cycle. An example of oncogenes would be the ras protein.
When proto-oncogenes’ activity is enhanced due to mutation, they become oncogenes and result in uncontrolled cell proliferation. Epigenetic changes can also increase the expression of proto-oncogenes. For instance, acetylation of the histone complexes around proto-oncogenes makes the DNA less tightly bound to the histone complex and more accessible by the transcription machinery. Increased expression of proto-oncogenes would have an oncogenic effect on the cell and result in uncontrolled cell division.
It is important to note that while mutations in TSGs are recessive, mutations in proto-oncogenes follow a dominant pattern. This means that for a cell to become cancerous and proliferate uncontrollably, both copies of TSGs would need to be mutated while mutation of only one of the proto-oncogene copies is enough to drive unregulated cell division.
Genomic imprinting can influence several human disorders, such as Prader-Willi syndrome (PWS) and Angelman syndrome (AS). Manifestations of PWS include impaired motor function, obesity, and tiny hands and feet. AS patients are skinny and energetic, have unique seizures and repeated symmetrical muscular movements, and suffer from impaired ability to acquire new information and knowledge.
The most prevalent cause of both PWS and AS is a deletion in the PWS and AS genes on human chromosome 15.
Only one copy of the chromosome 15 genes (maternal or paternal) is expressed in most individuals, while the other copy is kept inactive via epigenetic modifications. Therefore, most individuals possess one functional gene and one gene that has been epigenetically silenced.
If a mutation on chromosome 15 deletes the relevant genes, the offspring will inherit a set of non-functional genes and a set of epigenetically-inactive genes. This deletion leads to Angelman syndrome if inherited from the mother. But if the deletion is inherited from the father, it causes Prader-Willi syndrome.
Yes, epigenetics is a major part of biology. The word epigenetics essentially means ‘above genetics’ which describes factors separate to genetics. This is because epigenetic changes do not affect the base sequence of DNA.
Epigenetics is the study of mechanisms that lead to changes in gene expression that can be passed from cell to cell and are reversible but do not involve a change in the sequence of DNA
DNA methylation is an example of epigenetic modification which involves the addition of methyl groups to cystine bases in CpG islands in the DNA. This modification commonly inhibits the transcription of the genes.
An epigenetic trait is a characteristic which occurs in an organism due to the epigenetic changes occurring in their DNA. This characteristic occurs in the phenotype so an epigenetic trait is an observable characteristic that results from the interaction of the organism’s genotype with the environment i.e. epigenetics.
Environmental factors such as diet, physical activity, and psychological stress can cause an increase or decrease in epigenetic changes to DNA. There is much evidence proving lifestyle and environmental factors influence epigenetic mechanisms.
Flashcards in Epigenetics40
Start learningWhat is epigenetics?
Epigenetics is the study of mechanisms that lead to changes in gene expression that can be passed from cell to cell and are reversible, but do not involve a change in the sequence of DNA.
What are the three key types of epigenetic modifications that eukaryotes use to regulate gene expression?
DNA methylation, chromatin remodeling, and covalent histone modification.
What is the epigenome?
The epigenome is the set of all the chemical tags that are attached to the genome of a given cell.
What is DNA methylation?
DNA methylation is the process of addition of a methyl group to the cystine base in a CpG island on the DNA. The enzyme catalysing this process is DNA methyltransferase. This modification commonly inhibits the transcription of the genes.
What is histone acetylation?
During histone acetylation, an acetyl group is attached to a positively charged lysine residue in the amino-terminal tail of a histone core protein by histon acetyltransferase. This attachments eliminates the positive charge on the lysine side chain, hence it disrupts the electrostatic attraction between postively charged histones and negatively charged DNA strands. As a result of this, the DNA become less tightly bound to the histone proteins and become more accessible to RNA polymerase and TFs.
What are histones?
Histone complexes are structure that DNA double helix is wrapped around in nucleosomes.
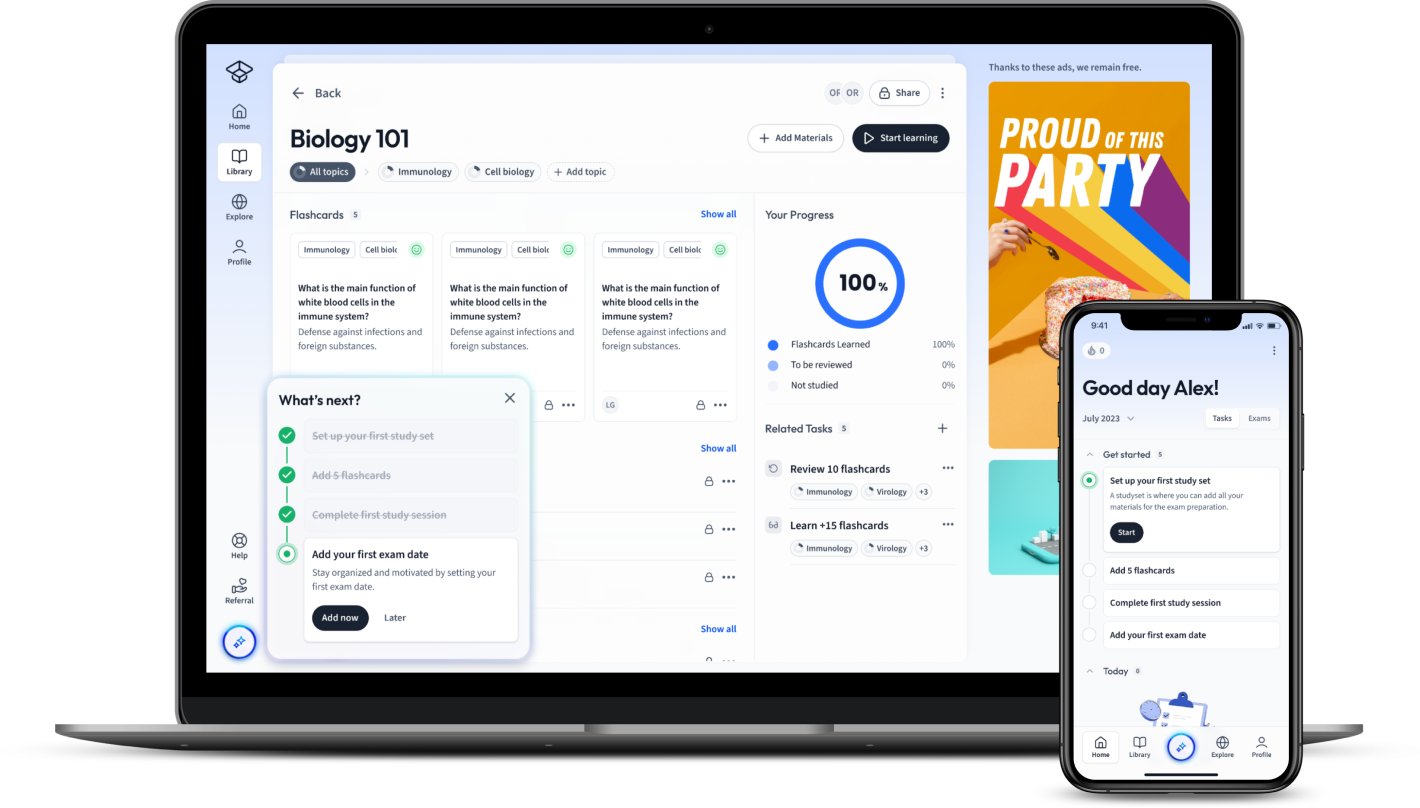
Already have an account? Log in
Open in AppThe first learning app that truly has everything you need to ace your exams in one place
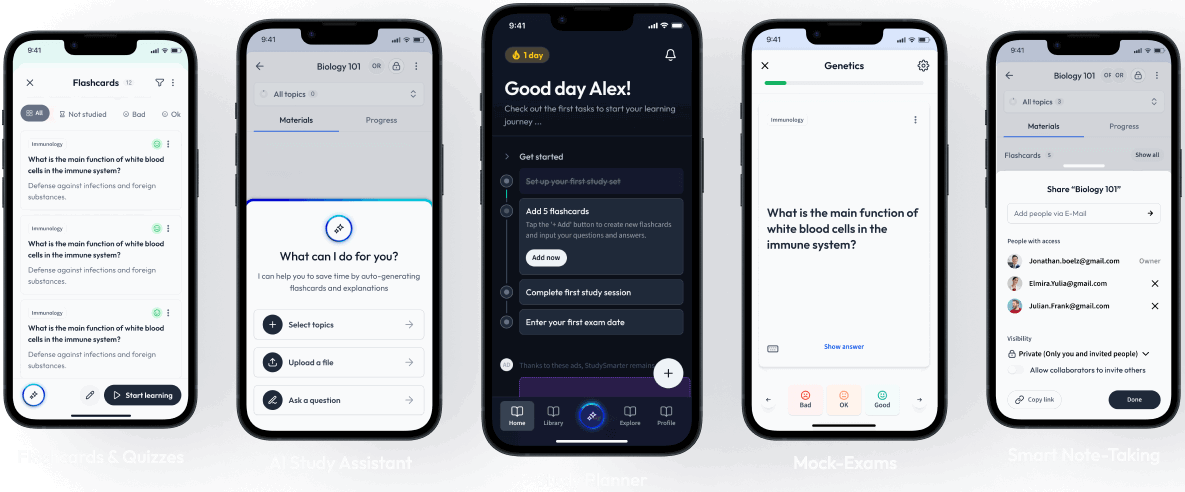
Sign up to highlight and take notes. It’s 100% free.
Save explanations to your personalised space and access them anytime, anywhere!
Sign up with Email Sign up with AppleBy signing up, you agree to the Terms and Conditions and the Privacy Policy of StudySmarter.
Already have an account? Log in